3 Chapter 3 exercises
Calculate \(\mathbf{A}^{\text{T}}\mathbf{B}\) in
Rwhere \(\mathbf{A}\) is a \(n \times p\) matrix comprising N(0, 1) random variates and \(\mathbf{B}\) is a \(n \times n\) matrix comprising Uniform([0, 1]) random variates for \(n = 1000\) and \(p = 500\), usingt(A) %*% Bandcrossprod(A, B).Confirm that both produce the same result.
Solution
## [1] TRUEThen benchmark the time that two commands take to complete.
Consider the calculation \(\mathbf{AD}\) where \[ \mathbf{A} = \begin{pmatrix}-0.42&-0.08&1.78&-0.08 \\-0.94&1.47&0.21&-0.84 \\0.95&0.04&1.78&-0.03 \\-0.09&-0.44&0.91&-2.07 \\-0.58&-0.83&-0.62&0.75 \\-0.94&0.09&0.13&0.65 \\\end{pmatrix} \text{ and }\mathbf{D} = \begin{pmatrix}-1.04&0.00&0.00&0.00 \\0.00&-0.41&0.00&0.00 \\0.00&0.00&-0.27&0.00 \\0.00&0.00&0.00&1.45 \\\end{pmatrix}. \] Write a function in
Rthat takes a matrixAand a vectordas its arguments and computes \(\mathbf{AD}\), where \(\mathbf{A} =\)Aand \(\text{diag}(\mathbf{D}) =\)d, and where \(\text{diag}(\mathbf{D})\) denotes the vector comprising the diagonal elements of \(\mathbf{D}\). Consider whether your function is performing redundant calculations and, if it is, try and avoid them.
Solution
We might start with
AD <- function(A, d) { # function to compute A * D # A is a matrix # D is a matrix with diagonal elements d # returns a matrix D <- diag(d) A %*% D }but if we do this then we’re multiplying a lot of zeros unnecessarily. Instead, the following avoids this
AdiagD <- function(A, d) { # function to compute A * D slightly more efficiently # A is a matrix # D is a matrix with diagonal elements d # returns a matrix t(t(A) * d) }and the following confirms that both give the same result
## [,1] [,2] [,3] [,4] ## [1,] 0.4368 0.0328 -0.4806 -0.1160 ## [2,] 0.9776 -0.6027 -0.0567 -1.2180 ## [3,] -0.9880 -0.0164 -0.4806 -0.0435 ## [4,] 0.0936 0.1804 -0.2457 -3.0015 ## [5,] 0.6032 0.3403 0.1674 1.0875 ## [6,] 0.9776 -0.0369 -0.0351 0.9425## [,1] [,2] [,3] [,4] ## [1,] 0.4368 0.0328 -0.4806 -0.1160 ## [2,] 0.9776 -0.6027 -0.0567 -1.2180 ## [3,] -0.9880 -0.0164 -0.4806 -0.0435 ## [4,] 0.0936 0.1804 -0.2457 -3.0015 ## [5,] 0.6032 0.3403 0.1674 1.0875 ## [6,] 0.9776 -0.0369 -0.0351 0.9425The following function generates an arbitrary \(n \times n\) positive definite matrix.
pdmatrix <- function(n) { # function to generate an arbitrary n x n positive definite matrix # n is an integer # returns a matrix L <- matrix(0, n, n) L[!lower.tri(L)] <- abs(rnorm(n * (n + 1) / 2)) tcrossprod(L) }By generating random \(n\)-vectors of independent N(0, 1) random variates, \(\mathbf{x}_1, \ldots, \mathbf{x}_m,\), say, and random \(n \times n\) positive definite matrices, \(\mathbf{A}_1, \ldots, \mathbf{A}_m\), say, confirm that \(\mathbf{x}_i^{\text{T}} \mathbf{A}_i \mathbf{x}_i > 0\) for \(i = 1, \ldots, m\) with \(m = 100\) and \(n = 10\).
[Note that this can be considered a simulation-based example of trying to prove a result by considering a large number of simulations. Such an approach can be very valuable when an analytical approach is not possible.]
Solution
There are a variety of ways we can tackle this. One of the tidier seems to be to use
all()andreplicate().check_pd <- function(A, x) { # function to check whether a matrix is positive definite # A is a matrix # returns a logical sum(x * (A %*% x)) > 0 } m <- 1e2 n <- 10 all(replicate(1e2, check_pd(pdmatrix(n), rnorm(n)) ))## [1] TRUEFor the cement factory data of Example 3.25 compute \(\hat {\boldsymbol \beta}\) by inverting \({\bf X}^{\text{T}} {\bf X}\) and multiplying by \({\bf X}^{\text{T}} {\bf y}\), i.e. \(\hat {\boldsymbol \beta} = ({\bf X}^{\text{T}} {\bf X})^{-1} {\bf X}^{\text{T}} {\bf y}\), and by solving \({\bf X}^{\text{T}} {\bf X} \hat {\boldsymbol \beta} = {\bf X}^{\text{T}} {\bf y}\).
Solution
X <- cbind(1, prod$days, prod$temp) XtX <- crossprod(X) y <- prod$output Xty <- crossprod(X, y) (betahat1 <- solve(XtX) %*% Xty)## [,1] ## [1,] 9.12688541 ## [2,] 0.20281539 ## [3,] -0.07239294## [,1] ## [1,] 9.12688541 ## [2,] 0.20281539 ## [3,] -0.07239294Show in
Rthat \(\mathbf{L}\) is a Cholesky decomposition of \(\mathbf{A}\) for \[ \mathbf{A} = \begin{pmatrix}547.56&348.66&306.54 \\348.66&278.26&199.69 \\306.54&199.69&660.38 \\\end{pmatrix} \text{ and }\mathbf{L} = \begin{pmatrix}23.4&0.0&0.0 \\14.9&7.5&0.0 \\13.1&0.6&22.1 \\\end{pmatrix}. \]
Solution
We’ll load \(\mathbf{A}\) and \(\mathbf{L}\) as
AandL, respectively.A <- matrix(c(547.56, 348.66, 306.54, 348.66, 278.26, 199.69, 306.54, 199.69, 660.38), 3, 3) L <- matrix(c(23.4, 14.9, 13.1, 0, 7.5, 0.6, 0, 0, 22.1), 3, 3)Then we need to show that \(\mathbf{L}\) is lower-triangular
## [1] TRUEwhich it is, that all its diagonal elements are positive
## [1] TRUEwhich they are, and that \(\mathbf{A} = \mathbf{LL}^{\text{T}}\)
## [1] TRUEwhich it does.
For the matrix \(\mathbf{A}\) below, find its Cholesky decomposition, \(\mathbf{L}\), where \(\mathbf{A} = \mathbf{LL}^\text{T}\) and \(\mathbf{L}\) is a lower triangular matrix, and confirm that \(\mathbf{L}\) is a Cholesky decomposition of \(\mathbf{A}\): \[\mathbf{A} = \begin{pmatrix}0.797&0.839&0.547 \\0.839&3.004&0.855 \\0.547&0.855&3.934 \\\end{pmatrix}.\]
Solution
We’ll load \(\mathbf{A}\)
and then we’ll find \(\mathbf{L}\)
## [,1] [,2] [,3] ## [1,] 0.8927486 0.0000000 0.000000 ## [2,] 0.9397943 1.4562921 0.000000 ## [3,] 0.6127145 0.1917022 1.876654Then we’ll check that it’s lower-triangular
## [1] TRUEwhich it is, then we’ll check that its diagonal elements are positive
## [1] TRUEwhich they are, and finally we’ll check that \(\mathbf{LL}^\text{T} = \mathbf{A}\)
## [1] TRUEwhich it does. So we conclude that \(\mathbf{L}\) is a lower-triangular Cholesky decomposition of \(\mathbf{A}\).
Form the matrices \[ \mathbf{A}_{11} = \left(\begin{array}{cc} 1 & 2\\ 3 & 4 \end{array}\right),~ \mathbf{A}_{12} = \left(\begin{array}{cc} 5 & 6\\ 7 & 8 \end{array}\right),~ \mathbf{A}_{21} = \left(\begin{array}{rr} 9 & 10\\ 11 & 12 \end{array}\right),~ \mathbf{A}_{22} = \left(\begin{array}{cc} 13 & 14\\ 15 & 16 \end{array}\right) \] in
Rand then use these to form \[ \mathbf{A} = \left(\begin{array}{cc} \mathbf{A}_{11} & \mathbf{A}_{12}\\ \mathbf{A}_{21} & \mathbf{A}_{22} \end{array}\right). \]
Solution
## [,1] [,2] ## [1,] 1 2 ## [2,] 3 4## [,1] [,2] ## [1,] 5 6 ## [2,] 7 8## [,1] [,2] ## [1,] 9 10 ## [2,] 11 12## [,1] [,2] ## [1,] 13 14 ## [2,] 15 16## [,1] [,2] [,3] [,4] ## [1,] 1 2 5 6 ## [2,] 3 4 7 8 ## [3,] 9 10 13 14 ## [4,] 11 12 15 16Using \(\mathbf{A}_{11}\) and \(\mathbf{A}_{12}\) from Question 7, compute \(\mathbf{A}_{11} \otimes \mathbf{A}_{12}\) in
R.
Solution
## [,1] [,2] [,3] [,4] ## [1,] 5 7 15 21 ## [2,] 6 8 18 24 ## [3,] 10 14 20 28 ## [4,] 12 16 24 32Repeat the formation of \(\mathbf{A}\) in Question 7 by considering a Kronecker sum.
Solution
One option is
## [,1] [,2] [,3] [,4] ## [1,] 1 3 5 7 ## [2,] 2 4 6 8 ## [3,] 9 11 13 15 ## [4,] 10 12 14 16but there are others.
Write a function in
Rcalledsolve_chol()to solve a system of linear equations \(\mathbf{Ax} = \mathbf{b}\) based on the Cholesky decomposition \(\mathbf{A} = \mathbf{LL}^{\text{T}}\).
Solution
The following is one option for
solve_chol().solve_chol <- function(L, b) { # Function to solve LL^Tx = b for x # L is a lower-triangular matrix # b is a vector of length nrow(L) # return vector of same length as b y <- forwardsolve(L, b) backsolve(t(L), y) }We’ll quickly check that we get the same result as
solve()using the data from Example 3.2.y <- c(.7, 1.3, 2.6) mu <- 1:3 Sigma <- matrix(c(4, 2, 1, 2, 3, 2, 1, 2, 2), 3, 3) res1 <- solve(Sigma, y - mu) L <- t(chol(Sigma)) all.equal(res1, solve_chol(L, y - mu))## [1] TRUENote above that we could use
backsolve(L, y, upper.tri = FALSE, transpose = TRUE)instead ofbacksolve(t(L), y), which avoids transposingL. Both give the same result, though.Show that solving \(\mathbf{Ax} = \mathbf{b}\) for \(\mathbf{x}\) is equivalent to solving \(\mathbf{Ly} = \mathbf{b}\) for \(\mathbf{y}\) and then \(\mathbf{L}^{\text{T}}\mathbf{x} = \mathbf{y}\) for \(\mathbf{x}\) if \(\mathbf{A}\) has Cholesky decomposition \(\mathbf{A} = \mathbf{L} \mathbf{L}^{\text{T}}\). Confirm this based on the cement factory data of Example 3.25 by taking \(\mathbf{A} = \mathbf{X}^{\text{T}}\mathbf{X}\), where \(\mathbf{X}\) is the linear model’s design matrix.
Solution
Let \(\mathbf{L}^{\text{T}} \mathbf{x} = \mathbf{y}\). To solve \(\mathbf{LL}^{\text{T}} \mathbf{x} = \mathbf{b}\) for \(\mathbf{x}\), we want to first solve \(\mathbf{Ly} = \mathbf{b}\) for \(\mathbf{y}\), and then \(\mathbf{L}^{\text{T}} \mathbf{x} = \mathbf{y}\) for \(\mathbf{x}\). We can confirm this numerically in
R.We already have
Xfrom Question \(\ref{cement}\), so we’ll re-use thatX. We’ll set \(\mathbf{A} = \mathbf{X}^{\text{T}} \mathbf{X}\), and call thisA. We’ll then usechol()to calculate its Cholesky decomposition in upper-triangular form,U, and lower-triangular form,L.The following two commands then solve \(\mathbf{Ly} = \mathbf{b}\) for \(\mathbf{y}\), and then \(\mathbf{L}^{\text{T}} \mathbf{x} = \mathbf{y}\) for \(\mathbf{x}\)
## [,1] [,2] ## [1,] 9.12688541 9.12688541 ## [2,] 0.20281539 0.20281539 ## [3,] -0.07239294 -0.07239294although double use of
solve()is inefficient compared to usingforwardsolve()and thenbacksolve().Alternatively, if we have \(\mathbf{A} = \mathbf{U}^{\text{T}} \mathbf{U}\), for upper-triangular \(\mathbf{U}\), then we have the following two options
cbind( backsolve(U, forwardsolve(t(U), Xty)), backsolve(U, forwardsolve(U, Xty, upper.tri = TRUE, transpose = TRUE)) )## [,1] [,2] ## [1,] 9.12688541 9.12688541 ## [2,] 0.20281539 0.20281539 ## [3,] -0.07239294 -0.07239294the latter of which is ever so slightly more efficient for its avoidance of
t(U).Show that \(\mathbf{U}\) and \(\boldsymbol{\Lambda}\) form an eigen-decomposition of \(\mathbf{A}\) for \[\mathbf{A} = \begin{pmatrix}3.40&0.00&0.00 \\0.00&0.15&-2.06 \\0.00&-2.06&-1.05 \\\end{pmatrix},~~\mathbf{U} = \begin{pmatrix}1.0&0.0&0.0 \\0.0&0.6&-0.8 \\0.0&0.8&0.6 \\\end{pmatrix} \text{ and } \boldsymbol{\Lambda} = \begin{pmatrix}3.4&0.0&0.0 \\0.0&-2.6&0.0 \\0.0&0.0&1.7 \\\end{pmatrix}.\]
Solution
We’ll load \(\mathbf{A}\), \(\boldsymbol{\Lambda}\) and \(\mathbf{U}\) as
A,LambdaandU, respectively.A <- cbind( c(3.4, 0, 0), c(0, .152, -2.064), c(0, -2.064, -1.052) ) Lambda <- diag(c(3.4, -2.6, 1.7)) U <- matrix(c(1, 0, 0, 0, .6, .8, 0, -.8, .6), 3, 3)Then we need to show that \(\mathbf{U}\) is orthogonal,
## [1] TRUEwhich it is, that \(\boldsymbol{\Lambda}\) is diagonal,
## [1] TRUEand that \(\mathbf{A} = \mathbf{U} \boldsymbol{\Lambda} \mathbf{U}^{\text{T}}\)
## [1] TRUEwhich it does.
For the matrix \(\mathbf{A}\) below, find \(\mathbf{U}\) and \(\boldsymbol{\Lambda}\) in its eigen-decomposition of the form \(\mathbf{A} = \mathbf{U} \boldsymbol{\Lambda} \mathbf{U}^\text{T}\)m where \(\mathbf{U}\) is orthogonal and \(\boldsymbol{\Lambda}\) is diagonal: \[\mathbf{A} = \begin{pmatrix}0.797&0.839&0.547 \\0.839&3.004&0.855 \\0.547&0.855&3.934 \\\end{pmatrix}.\]
Solution
We’ll load \(\mathbf{A}\)
and then find \(\mathbf{U}\) and \(\boldsymbol{\Lambda}\)
## [,1] [,2] [,3] ## [1,] -0.2317139 -0.1941590 0.95321085 ## [2,] -0.5372074 -0.7913728 -0.29178287 ## [3,] -0.8109975 0.5796821 -0.07906848## [,1] [,2] [,3] ## [1,] 4.656641 0.000000 0.0000000 ## [2,] 0.000000 2.583555 0.0000000 ## [3,] 0.000000 0.000000 0.4948042Then we need to show that \(\mathbf{U}\) is orthogonal,
## [1] TRUEwhich it is, that \(\boldsymbol{\Lambda}\) is diagonal,
## [1] TRUEwhich it is, and that \(\mathbf{A} = \mathbf{U} \boldsymbol{\Lambda} \mathbf{U}^{\text{T}}\)
## [1] TRUEwhich it does.
Show that \(\mathbf{U}\), \(\mathbf{D}\) and \(\mathbf{V}\) form a singular value decomposition of \(\mathbf{A}\) for
\[\mathbf{A} = \begin{pmatrix}0.185&8.700&-0.553 \\0.555&-2.900&-1.659 \\3.615&8.700&-2.697 \\1.205&-26.100&-0.899 \\\end{pmatrix},~~\mathbf{U} = \begin{pmatrix}-0.2&-0.2&1.0 \\-0.5&-0.8&-0.3 \\-0.8&0.6&-0.1 \\\end{pmatrix},\] \[\mathbf{D} = \begin{pmatrix}29&0&0 \\0&5&0 \\0&0&1 \\\end{pmatrix} \text{ and } \mathbf{V} = \begin{pmatrix}0.00&0.76&0.65 \\-1.00&0.00&0.00 \\0.00&-0.65&0.76 \\\end{pmatrix}.\]
Solution
We’ll load \(\mathbf{A}\), \(\mathbf{U}\), \(\mathbf{D}\) and \(\mathbf{V}\) as
A,U,DandV, respectively.A <- matrix(c(0.185, 0.555, 3.615, 1.205, 8.7, -2.9, 8.7, -26.1, -0.553, -1.659, -2.697, -0.899), 4, 3) U <- matrix(c(-0.3, 0.1, -0.3, 0.9, 0.1, 0.3, 0.9, 0.3, -0.3, -0.9, 0.3, 0.1), 4, 3) D <- diag(c(29, 5, 1)) V <- matrix(c(0, -1, 0, 0.76, 0, -0.65, 0.65, 0, 0.76), 3, 3)We want to check that \(\mathbf{U}\) and \(\mathbf{V}\) are orthogonal
## [1] TRUE## [1] "Mean relative difference: 9.999e-05"which they both are, that \(\mathbf{D}\) is diagonal
## [1] TRUE## [1] "Mean relative difference: 9.999e-05"which it is, and finally that \(\mathbf{A} = \mathbf{UDV}^{\text{T}}\)
## [1] TRUEwhich is true.
By considering \(\sqrt{\mathbf{A}}\) as \(\mathbf{A}^{1/2}\), i.e. as a matrix power, show how an eigen-decomposition can be used to general multivariate Normal random vectors and then write a function to implement this in
R.
Solution
From Example 3.14, to generate a multivariate Normal random vector, \(\mathbf{Y}\), say, from the \(MVN_p({\boldsymbol \mu}, {\boldsymbol \Sigma})\) distribution we need to compute \(\mathbf{Y} = \boldsymbol{\mu} + \mathbf{L} \mathbf{Z}\), where \(\boldsymbol{\Sigma} = \mathbf{L} \mathbf{L}^{\text{T}}\) and \(\mathbf{Z} = (Z_1, \ldots, Z_p)^{\text{T}}\), where \(Z_i\), \(i = 1, \ldots, p\), are independent \(N(0, 1)\) random variables. Given an eigen-decomposition of \({\boldsymbol \Sigma} = \mathbf{U} \boldsymbol{\Lambda} \mathbf{U}^{\text{T}}\), we can write this as \({\boldsymbol \Sigma} = \mathbf{U} \boldsymbol{\Lambda}^{1/2} \boldsymbol{\Lambda}^{1/2} \mathbf{U}^{\text{T}}\). As \(\boldsymbol{\Lambda}\) is diagonal, \(\boldsymbol{\Lambda} = \boldsymbol{\Lambda}^{\text{T}}\) and hence \(\boldsymbol{\Lambda}^{1/2} \mathbf{U}^{\text{T}} = (\mathbf{U} \boldsymbol{\Lambda}^{1/2})^{\text{T}}\) and so \({\boldsymbol \Sigma} = \mathbf{LL}^{\text{T}}\) if \(\mathbf{L} = \mathbf{U} \boldsymbol{\Lambda}^{1/2}\). Therefore we can generate \(MVN_p({\boldsymbol \mu}, {\boldsymbol \Sigma})\) random variables with \(\mathbf{Y} = \boldsymbol{\mu} + \mathbf{U} \boldsymbol{\Lambda}^{\text{1/2}} \mathbf{Z}\).
The following
Rfunction generatesnmultivariate Normal random vectors with meanmuand variance-covariance matrixSigma.rmvn_eigen <- function(n, mu, Sigma) { # Function to generate MVN random vectors with # n is a integer, giving the number of independent vectors to simulate # mu is a p-vector of the MVN mean # Sigma is a p x p matrix of the MVN variance-covariance matrix # returns a p times n matrix eS <- eigen(Sigma, symmetric = TRUE) p <- nrow(Sigma) Z <- matrix(rnorm(p * n), p) mu + eS$vectors %*% diag(sqrt(eS$values)) %*% Z } rmvn_eigen(5, mu, Sigma)## [,1] [,2] [,3] [,4] [,5] ## [1,] 3.812779 1.328476 4.819841 -1.4787812 -0.6061167 ## [2,] 5.302633 3.159313 3.723752 -0.1035905 -0.5120484 ## [3,] 5.310861 4.700222 3.865721 2.8249234 0.7614778Show how an eigen-decomposition can be used to solve a system of linear equations \(\mathbf{Ax} = \mathbf{b}\) for \(\mathbf{x}\) by matrix multiplications and vector divisions, only. Confirm this in
Rby solving \(\boldsymbol{\Sigma} \mathbf{z} = \mathbf{y} - \boldsymbol{\mu}\) for \(\mathbf{z}\), with \(\mathbf{y}\), \(\boldsymbol{\mu}\) and \(\boldsymbol{\Sigma}\) as in Example 3.2.
Solution
To solve \(\mathbf{Ax} = \mathbf{b}\) for \(\mathbf{x}\), if \(\mathbf{A} = \mathbf{U} \boldsymbol{\Lambda} \mathbf{U}^{\text{T}}\), then we want to solve \(\mathbf{U} \boldsymbol{\Lambda} \mathbf{U}^{\text{T}} \mathbf{x} = \mathbf{b}\) for \(\mathbf{x}\). The following manipulations can be used \[\begin{align} \boldsymbol{\Lambda} \mathbf{U}^{\text{T}} \mathbf{x} &= \mathbf{U}^{-1} \mathbf{b} \tag{3.1}\\ \boldsymbol{\Lambda} \mathbf{U}^{\text{T}} \mathbf{x} &= \mathbf{U}^{\text{T}} \mathbf{b} \tag{3.2} \\ \mathbf{U}^{\text{T}} \mathbf{x} &= (\mathbf{U}^{\text{T}} \mathbf{b}) / \text{diag}(\boldsymbol{\Lambda}) \tag{3.3} \\ \mathbf{x} &= \mathbf{U}^{-\text{T}}(\mathbf{U}^{\text{T}} \mathbf{b}) / \text{diag}(\boldsymbol{\Lambda}) \tag{3.4} \\ \mathbf{x} &= \mathbf{U} [(\mathbf{U}^{\text{T}} \mathbf{b}) / \text{diag}(\boldsymbol{\Lambda})] \tag{3.5} \end{align}\] in which (3.1) results from premultiplying by \(\mathbf{U}^{\text{-1}}\), (3.2) from orthogonality of \(\mathbf{U}\), i.e. \(\mathbf{U}^{\text{T}} = \mathbf{U}^{-1}\), (3.3) from elementwise division, given diagonal \(\boldsymbol{\Lambda}\), (3.4) from premultiplying by \(\mathbf{U}^{-\text{T}}\) and (3.5) from orthogonality of \(\mathbf{U}\), again.
Next we’ll load the data from Example 3.2
y <- c(.7, 1.3, 2.6) mu <- 1:3 Sigma <- matrix(c(4, 2, 1, 2, 3, 2, 1, 2, 2), 3, 3) res1 <- solve(Sigma, y - mu)Then we’ll go through the calculations given above
eS <- eigen(Sigma, symmetric = TRUE) lambda <- eS$values U <- eS$vectors res2 <- U %*% (crossprod(U, y - mu) / lambda)which we see gives the same result as
solve(), once we useas.vector()to convertres2from a one-column matrix to a vector.## [1] TRUE
Show in
Rthat if \(\mathbf{H}_{4} = \mathbf{UDV}^{\text{T}}\) is the SVD of \(\mathbf{H}_4\), the \(4 \times 4\) Hilbert matrix, then solving \(\mathbf{H}_4\mathbf{x} = (1, 1, 1, 1)^{\text{T}}\) for \(\mathbf{x}\) reduces to solving \[ \mathbf{V}^{\text{T}} \mathbf{x} \simeq \begin{pmatrix}-1.21257 \\-4.80104 \\-26.08668 \\234.33089 \\\end{pmatrix} \] and then use this to solve \(\mathbf{H}_4\mathbf{x} = (1, 1, 1, 1)^{\text{T}}\) for \(\mathbf{x}\).
Solution
We ultimately need to solve \(\mathbf{UDV}^{\text{T}} \mathbf{x} = \mathbf{b}\), where \(\mathbf{b} = (1, 1, 1, 1)^{\text{T}}\). Pre-multiplying by \(\mathbf{U}^{-1} = \mathbf{U}^{\text{T}}\), we then need to solve \(\mathbf{DV}^{\text{T}} \mathbf{x} = \mathbf{U}^{\text{T}} \mathbf{b}\), which is equivalent to solving \(\mathbf{V}^{\text{T}} \mathbf{x} = (\mathbf{U}^{\text{T}} \mathbf{b}) / \text{diag}(\mathbf{D})\). The following calculates the SVD of \(\mathbf{H}_4\) and then computes \((\mathbf{U}^{\text{T}} \mathbf{b}) / \text{diag}(\mathbf{D})\).
hilbert <- function(n) { # Function to evaluate n by n Hilbert matrix. # n is an integer # Returns n by n matrix. ind <- 1:n 1 / (outer(ind, ind, FUN = '+') - 1) } H4 <- hilbert(4) b <- rep(1, 4) svdH <- svd(H4) V <- svdH$v U <- svdH$u b2 <- crossprod(U, b) z <- b2 / svdH$d z## [,1] ## [1,] -1.212566 ## [2,] -4.801038 ## [3,] -26.086677 ## [4,] 234.330888which is as given in the question, subject to rounding. Finally we want to solve \(\mathbf{V}^{\text{T}} \mathbf{x} = (\mathbf{U}^{\text{T}} \mathbf{b}) / \text{diag}(\mathbf{D})\) for \(\mathbf{x}\), which we can do with either of the following
## [,1] [,2] ## [1,] -4 -4 ## [2,] 60 60 ## [3,] -180 -180 ## [4,] 140 140and we see that the latter gives the same result as
solve()## [1] TRUEonce we ensure that both are vectors. Note that solving systems of linear equations via the SVD is only a sensible option if we already have the SVD. Otherwise, solving via other decompositions is more efficient.
Show that \(\mathbf{Q}\) and \(\mathbf{R}\) form a QR decomposition of \(\mathbf{A}\) for \[\mathbf{A} = \begin{pmatrix}0.337&0.890&-1.035 \\0.889&6.070&-1.547 \\-1.028&-1.545&4.723 \\\end{pmatrix},~~\mathbf{Q} = \begin{pmatrix}-0.241&0.101&0.965 \\-0.635&-0.769&-0.078 \\0.734&-0.631&0.249 \\\end{pmatrix}\] and \[\mathbf{R} = \begin{pmatrix}-1.4&-5.2&4.7 \\0.0&-3.6&-1.9 \\0.0&0.0&0.3 \\\end{pmatrix}.\]
Solution
We’ll load \(\mathbf{A}\), \(\mathbf{Q}\) and \(\mathbf{R}\) as
A,QandR, respectively.A <- matrix(c(0.3374, 0.889, -1.0276, 0.8896, 6.0704, -1.5452, -1.0351, -1.5468, 4.7234), 3, 3) Q <- matrix(c(-0.241, -0.635, 0.734, 0.101, -0.769, -0.631, 0.965, -0.078, 0.249), 3, 3) R <- matrix(c(-1.4, 0, 0, -5.2, -3.6, 0, 4.7, -1.9, 0.3), 3, 3)We want to show that \(\mathbf{Q}\) is orthogonal
## [1] "Mean relative difference: 0.001286839"which it is (after allowing for a bit of error), that \(\mathbf{R}\) is upper-triangular
## [1] TRUEwhich it is, and that \(\mathbf{A} = \mathbf{QR}\)
## [1] TRUEwhich it does.
The determinant of the \(n \times n\) Hilbert matrix is given by \[ |\mathbf{H}_n| = \dfrac{c_n^4}{c_{2n}} \] where \[ c_n = \prod_{i = 1}^{n - 1} i! \] is the Cauchy determinant.
Write a function in
R,det_hilbert(n, log = FALSE)that evaluates \(|\mathbf{H}_n|\) and \(\log(|\mathbf{H}_n|)\) iflog = FALSEorlog = TRUE, respectively. Your function should compute \(\log(|\mathbf{H}_n|)\), and then return \(|\mathbf{H}_n|\) iflog = FALSE, as withdmvn1()in Example 3.2.
Solution
It will perhaps be tidier to write a function to calculate the Cauchy determinant
det_cauchy <- function(n, log = FALSE) { # function to calculate Cauchy determinant # n in an integer # log is a logical; defaults to FALSE # returns a scalar out <- sum(lfactorial(seq_len(n - 1))) if (!log) out <- exp(out) out }and then to use that to calculate the determinant of \(\mathbf{H}_n\).
Calculate \(|\mathbf{H}_n|\) and \(\log(|\mathbf{H}_n|)\) through the QR decomposition of \(\mathbf{H}_n\) for \(n = 5\) and confirm that both give the same result as
det_hilbert()above.
Solution
We’ll start with a function
det_QR(), which calculates the determinant of a matrix via its QR decompositiondet_QR <- function(A) { # function to calculate determinant of a matrix via QR decomposition # A is a matrix # returns a scalar qrA <- qr(A) R <- qr.R(qrA) prod(abs(diag(R))) }and we see that this gives the same answer as
det_hilbert()for \(n = 5\).## [1] 3.749295e-12## [1] 3.749295e-12Then we’ll write a function to calculate the logarithm of the determinant of a matrix through its QR decomposition.
logdet_QR <- function(A) { # function to calculate log determinant of a matrix via QR decomposition # A is a matrix # returns a scalar qrA <- qr(A) R <- qr.R(qrA) sum(log(abs(diag(R)))) }which we also see gives the same answer as
det_hilbert()withlog = TRUE.## [1] -26.30945## [1] -26.30945
Compute \(\mathbf{H}_4^{-1}\) via its QR decomposition, and confirm your result with
solve()andqr.solve().
Solution
If \(\mathbf{H}_4 = \mathbf{QR}\) then \(\mathbf{H}_4^{-1} = (\mathbf{QR})^{-1} = \mathbf{R}^{-1} \mathbf{Q}^{-1} = \mathbf{R}^{-1} \mathbf{Q}^{\text{T}}\), since \(\mathbf{Q}\) is orthogonal. Therefore we want to solve \(\mathbf{R} \mathbf{X} = \mathbf{Q}^{\text{T}}\) for \(\mathbf{X}\). The following calculates the QR decomposition of \(\mathbf{H}_4\) with
qr()and then extracts \(\mathbf{Q}\) and \(\mathbf{R}\) asQandR, respectively.Then we want to solve \(\mathbf{R} \mathbf{X} = \mathbf{Q}^{\text{T}}\) for \(\mathbf{X}\), which we do with
backsolve(), since \(\mathbf{R}\), stored asR, is upper-triangular.The following use
solve()andqr.solve(). Note that if we already have the QR decomposition fromqr(), thenqr.solve()uses far fewer calculations to obtain the inverse.We see that both give the same answer
## [1] TRUEand also the same answer as with
backsolve()above## [1] TRUEBenchmark Cholesky, eigen (with
symmetric = TRUEandsymmetric = FALSE), singular value and QR decompositions of \(\mathbf{A} = \mathbf{I}_{100} + \mathbf{H}_{100}\), where \(\mathbf{H}_{100}\) is the \(100 \times 100\) Hilbert matrix. (If you’re feeling impatient, consider reducing the value of argumenttimesfor functionmicrobenchmark::microbenchmark().)
Solution
hilbert <- function(n) { # Function to evaluate n by n Hilbert matrix. # n is an integer # Returns n by n matrix. ind <- 1:n 1 / (outer(ind, ind, FUN = '+') - 1) }H100 <- hilbert(1e2) + diag(1, 1e2) microbenchmark::microbenchmark( chol(H100), eigen(H100, symmetric = TRUE), eigen(H100), svd(H100), qr(H100), times = 1e2 )## Unit: microsecondsIf we consider median computation times, we see that the Cholesky decomposition is quickest, at nearly six times quicker than the QR decomposition, which is next quickest. The QR decomposition is then about just under three times quicker than the symmetric eigen-decomposition, which takes about the same amount of time as the singular value decomposition. The asymmetric eigen-decomposition is slowest, and demonstrates that if we know the matrix we want an eigen-decomposition of is symmetric, then we should pass this information to
R.Remark. The following shows us that if we only want the eigenvalues of a symmetric matrix, then we can further save times by specifying
only.values = TRUE.microbenchmark::microbenchmark( eigen(H100, symmetric = TRUE), eigen(H100, symmetric = TRUE, only.values = TRUE), times = 1e2 )## Unit: millisecondsGiven that \[\mathbf{A} = \begin{pmatrix}1.23&0.30&2.58 \\0.30&0.43&1.92 \\2.58&1.92&10.33 \\\end{pmatrix},~\mathbf{A}^{-1} = \begin{pmatrix}6.9012&16.9412&-4.8724 \\16.9412&55.2602&-14.5022 \\-4.8724&-14.5022&4.0092 \\\end{pmatrix},\] \[\mathbf{B} = \begin{pmatrix}1.49&0.40&2.76 \\0.40&0.53&2.16 \\2.76&2.16&11.20 \\\end{pmatrix},\] and that \(\mathbf{B} = \mathbf{A} + \mathbf{LL}^{\text{T}}\), where \(\mathbf{L}\) is a lower-triangular matrix, find \(\mathbf{B}^{-1}\) using Woodbury’s formula, i.e. using \[(\mathbf{A} + \mathbf{UV}^{\text{T}})^{-1} = \mathbf{A}^{-1} - \mathbf{A}^{-1} \mathbf{U} (\mathbf{I}_n + \mathbf{V}^{\text{T}} \mathbf{A}^{-1} \mathbf{U})^{-1} \mathbf{V}^{\text{T}} \mathbf{A}^{-1}. \]
Solution
We note that we can write \(\mathbf{U} = \mathbf{V} = \mathbf{L}\) and that \(\mathbf{LL}^{\text{T}} = \mathbf{B} - \mathbf{A}\), and so we can obtain \(\mathbf{L}\) via the Cholesky decomposition of \(\mathbf{B} - \mathbf{A}\). We’ll load \(\mathbf{A}\), \(\mathbf{A}^{-1}\) and \(\mathbf{B}\) below as
A,iAandB, and then compute the lower-triangular Cholesky decomposition of \(\mathbf{B} - \mathbf{A}\) and store this asL.A <- matrix( c(1.23, 0.3, 2.58, 0.3, 0.43, 1.92, 2.58, 1.92, 10.33), 3, 3) iA <- matrix( c(6.9012, 16.9412, -4.8724, 16.9412, 55.2602, -14.5022, -4.8724, -14.5022, 4.0092), 3, 3) B <- matrix( c(1.49, 0.4, 2.76, 0.4, 0.53, 2.16, 2.76, 2.16, 11.2), 3, 3) L <- t(chol(B - A))Then we’ll write a function to implement Woodbury’s formula,
woodbury <- function(iA, U, V) { # function to implement Woodbury's formula # iA, U and V are matrices # returns a matrix I_n <- diag(nrow(iA)) iAU <- iA %*% U iA - iAU %*% solve(I_n + crossprod(V, iAU), crossprod(V, iA)) }and then use this to find \(\mathbf{B}^{-1}\), which we’ll call
iB,and see that, subject to rounding, this is equal to \(\mathbf{B}^{-1}\)
## [1] "Mean relative difference: 9.113679e-06"Recall the cement factory data of Example 3.25. Now suppose that a further observation has been obtained based on the factory operating at 20 degrees for 14 days. For given \(\sigma^2\), the sampling distribution of \(\hat{\boldsymbol{\beta}}\) is \(MVN_3(\hat{\boldsymbol{\beta}}, \sigma^{-2} (\mathbf{X}^{\text{T}}\mathbf{X})^{-1})\). Use the Sherman-Morrison formula to give an expression for the estimated standard errors of \(\hat{\boldsymbol{\beta}}\) in terms of \(\sigma\) given that
\[(\mathbf{X}^{\text{T}}\mathbf{X})^{-1} = \begin{pmatrix} 2.78 \times 10^0 & -1.12 \times 10^{-2} & -1.06 \times 10^{-1}\\ -1.12 \times 10^{-2} & 1.46 \times 10^{-4} & 1.75 \times 10^{-4}\\ -1.0 \times 10^{-1} & 1.75 \times 10^{-4} & 4.79 \times 10^{-3} \end{pmatrix}. \]
Solution
If we refer to the Sherman-Morrison formula, we take \(\mathbf{A}^{-1} = (\mathbf{X}^{\text{T}}\mathbf{X})^{-1}\) and \(\mathbf{u} = \mathbf{v} = (1, 14, 20)^{\text{T}}\). Then we can calculate the updated variance-covariance matrix of the sampling distribution of \(\hat{\boldsymbol{\beta}}\) as
\[\sigma^{-2} (\mathbf{A} + \mathbf{u} \mathbf{v}^{\text{T}})^{-1} = \sigma^{-1} \left[ \mathbf{A}^{-1} - \dfrac{\mathbf{A}^{-1} \mathbf{u} \mathbf{v}^{\text{T}} \mathbf{A}^{-1}}{1 + \mathbf{v}^{\text{T}}\mathbf{A}^{-1}\mathbf{u}} \right] \]
which can be calculated in
RwithV <- matrix( c(2.78, -0.0112, -0.106, -0.0112, 0.000146, 0.000175, -0.106, 0.000175, 0.00479), 3, 3) u <- c(1, 14, 20) V2 <- V - (V %*% tcrossprod(u) %*% V) / (1 + crossprod(u, V %*% u))[1, 1] V2## [,1] [,2] [,3] ## [1,] 2.580467260 -0.0089572393 -0.1029269103 ## [2,] -0.008957239 0.0001207912 0.0001404583 ## [3,] -0.102926910 0.0001404583 0.0047426700Taking the diagonal elements of
V2we get\[ \left(\text{e.s.e.}(\hat \beta_0), \text{e.s.e.}(\hat \beta_1), \text{e.s.e.}(\hat \beta_2)\right) = \sigma^{-1} \left( 1.606~,0.011~,0.069 \right). \]
Consider a polynomial regression model of the form
\[ Y \mid x \sim N(m(x), \sigma^2) \]
for \(\sigma > 0\) and with
\[ m(x) = \beta_0 + \sum_{i = 1}^p \beta_i x^i \]
for \((p + 1)\)-vector of regression coefficients \(\boldsymbol{\beta} = (\beta_0, \beta_1, \dots, \beta_p)^\textsf{T}\).
Now let \(Y_i\), where \(i = 1, 2, \ldots, 145\), denote the global surface temperature anomaly for years \(1880, 1881, \ldots, 2024\). Then let \(x_i = i - 70\). We can load these data in
RwithFor \(p = 3\), estimate \(E(Y_i \mid x_i)\) for \(i = 1, \ldots, 145\) using maximum likelihood.
Solution
Note that we can write \(E(Y \mid x) = \mathbf{x}^\textsf{T} \boldsymbol{\beta}\), where \(\mathbf{x}^\textsf{T} = c(1, x, x^2, \ldots, x^p)\). Then our maximum likelihood estimate of \(\boldsymbol{\beta}\) is \(\hat{\boldsymbol{\beta}} = (\mathbf{X}^\textsf{T} \mathbf{X})^{-1} \mathbf{X}^\textsf{T} \mathbf{y}\), where \(\mathbf{X}\) has \(i\)th row \(\mathbf{x}_i^\textsf{T} = (1, x_i, x_i^2, x_i^3)\). We can form this with
and then read in the response data with
Then we can obtain \(\hat{\boldsymbol{\beta}}\) with
Finally, our estimates of \(E(Y_1 \mid x_1), \ldots, E(Y_{145} \mid x_{145})\) can be calculated as \(\hat{ \mathbf{y}} = \mathbf{X} \hat{\boldsymbol{\beta}}\), as below
(Aside: these are the data seen in the so-called hockey stick graph.
plot(data$year, data$temperature, xlab = 'Year', ylab = 'Global surface temperature anomaly', main = 'Global surface temperature anomaly by year', pch = 19) lines(data$year, y_hat, col = 2, lwd = 3) legend('topleft', legend = c('Data', 'Polynomial fit'), pch = c(19, -1), lwd = c(0, 3), col = c(1, 2))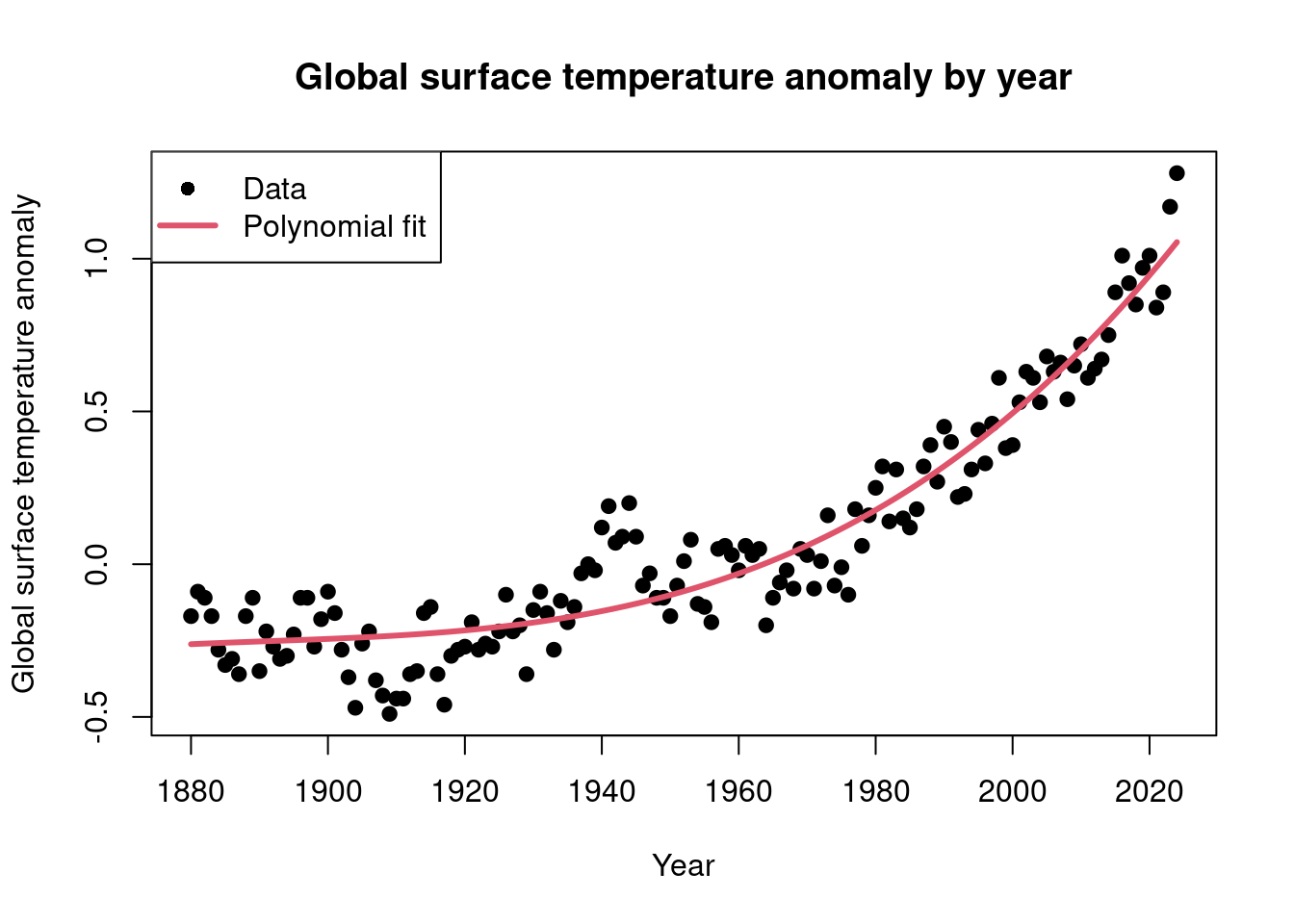
Sometimes, the data are plotted as blues and reds
image(x = data$year, z = matrix(data$temperature, ncol = 1), col = hcl.colors(11, 'Blue-Red 3'), xlab = 'Year', ylab = '', main = 'Color-based global surface temperature anomaly by year')
as in the plot above.)